Juiceflow - GCP
Introduction
Juiceflow combines JuiceFS and Nextflow on MMCloud, offering a powerful, scalable solution for managing and executing workflows in the cloud.
Expand for more details on JuiceFS
JuiceFS is an open-source, high-performance distributed file system designed specifically for cloud environments. It offers unique features, such as:
- Separation of Data and Metadata: JuiceFS stores files in chunks within object storage like Google Cloud Storage, while metadata can be stored in various databases, including Redis.
- Performance: Achieves millisecond-level latency and nearly unlimited throughput, depending on the object storage scale.
- Easy Integration with MMCloud: MMCloud provides pre-configured nextflow head node templates with JuiceFS setup, simplifying deployment.
- Comparison with S3FS: For a detailed comparison between JuiceFS and S3FS, see JuiceFS vs. S3FS. JuiceFS typically offers better performance and scalability.
Pre-requisites
-
GCP
gs://buckets with proper permissions to your service account. -
GCP Credential key for a service account. To create one navigate to Google Cloud Console
-
Navigate to
IAM->Service Accounts, click your corresponding account ->KEYStab.
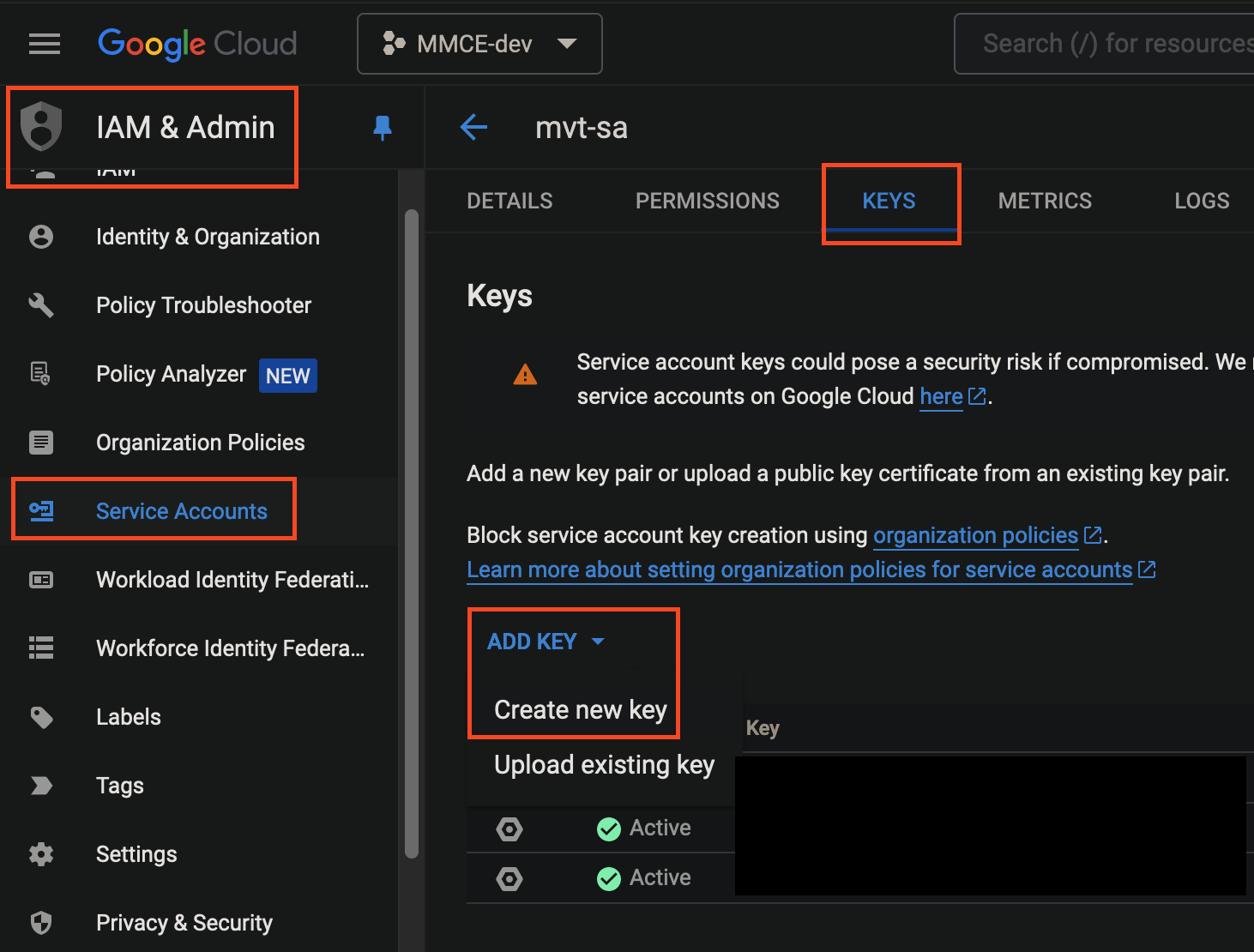
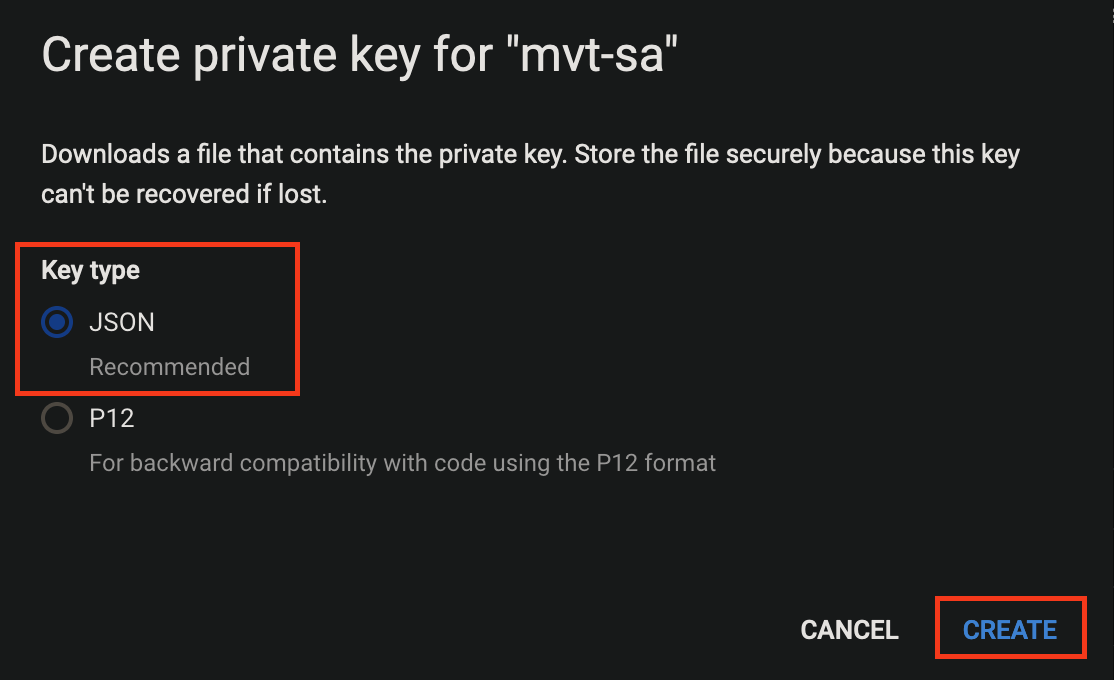
- Create a JSON key and the key will be downloaded to your local machine upon creation. This key will be set as float secret and used later.
Overview of the Setup
This solution leverages two scripts:
transient_JFS_GCP.sh: Formats the work directory bucket to JuiceFS format.job_submit_GCP.sh: Contains Nextflow input parameters and MMC config.
Steps
Download Scripts
- Host-init script (you don't have to edit this script, but you'll need it later)
wget https://mmce-data.s3.amazonaws.com/juiceflow/v1/gcp/transient_JFS_GCP.sh- Job-submit script (Download the template or create one locally based on the template below with your Nextflow inputs and run configurations)
wget https://mmce-data.s3.amazonaws.com/juiceflow/v1/gcp/job_submit_GCP.shExpand to view a sample job-submit.sh script
#!/bin/bash
# ---- User Configuration Section ----
# These configurations must be set by the user before running the script.
# ---- Optional Configuration Section ----
# These configurations are optional and can be customized as needed.
# JFS (JuiceFS) Private IP: Retrieved from the WORKER_ADDR environment variable.
jfs_private_ip=$(echo $WORKER_ADDR)
# Work Directory: Defines the root directory for working files. Optional suffix can be added.
workDir_suffix=''
workDir='/mnt/jfs/'$workDir_suffix
mkdir -p $workDir # Ensures the working directory exists.
cd $workDir # Changes to the working directory.
export NXF_HOME=$workDir # Sets the NXF_HOME environment variable to the working directory.
# ---- Nextflow Configuration File Creation ----
# This section creates a Nextflow configuration file with various settings for the pipeline execution.
# Use cat to create or overwrite the mmc.config file with the desired Nextflow configurations.
# NOTE: GCS keys and OpCenter information will be concatted to the end of the config file. No need to add them now
cat > mmc.config << EOF
// enable nf-float plugin.
plugins {
id 'nf-float'
}
// Process settings: Executor, error strategy, and resource allocation specifics.
process {
executor = 'float'
errorStrategy = 'retry'
extra = '--dataVolume [opts=" --cache-dir /mnt/jfs_cache "]jfs://${jfs_private_ip}:6868/1:/mnt/jfs --dataVolume [size=120]:/mnt/jfs_cache --vmPolicy [spotOnly=true,retryLimit=10,retryInterval=300s]'
}
// Directories for Nextflow execution.
workDir = '${workDir}'
launchDir = '${workDir}'
EOF
# ---- Data Preparation ----
# Use this section to copy essential files from GCS to the working directory.
# For example, copy the sample sheet and params.yml from GCS to the current working directory.
# gcloud storage gs://nextflow-input/samplesheet.csv .
# gcloud storage gs://nextflow-input/scripts/params.yml .
# ---- Nextflow Command Setup ----
# Important: The -c option appends the mmc config file and soft overrides the nextflow configuration.
# Assembles the Nextflow command with all necessary options and parameters.
# The gs path needs to contain at least a sub-directory
# Bucket name MUST NOT contain underscores due to an internal GCS limitation
nextflow_command='nextflow run <nextflow-pipeline> \
-r <revision-number> \
-c mmc.config \
-params-file params.yml \
--input samplesheet.csv \
--outdir 'gs://nextflow-output/rnaseq/' \
-resume '
# -------------------------------------
# ---- DO NOT EDIT BELOW THIS LINE ----
# -------------------------------------
# The following section contains functions and commands that should not be modified by the user.
function install_float {
# Install float
local address=$(echo "$FLOAT_ADDR" | cut -d':' -f1)
wget https://$address/float --no-check-certificate --quiet
chmod +x float
}
function get_secret {
input_string=$1
local address=$(echo "$FLOAT_ADDR" | cut -d':' -f1)
secret_value=$(./float secret get $input_string -a $address)
if [[ $? -eq 0 ]]; then
# Have this secret, will use the secret value
echo $secret_value
return
else
# Don't have this secret, will still use the input string
echo $1
fi
}
function gcloud_init() {
# Login to gcloud with credential key
gcloud auth login --quiet --cred-file=/mnt/jfs/gcp_cred.json
if [[ $? -ne 0 ]]; then
echo $(date): "gcloud authentication failed. Cannot dump JuiceFS metadata."
exit 1
fi
}
function remove_old_metadata () {
echo $(date): "First finding and removing old metadata..."
FOUND_METADATA=$(gcloud storage ls $BUCKET | grep ".meta.json.gz" |awk '{print $1}' | sed 's/gs:\/\///' | cut -d'/' -f2-)
if [[ -z "$FOUND_METADATA" ]]; then
echo $(date): "No previous metadata dump found. Continuing with dumping current JuiceFs"
else
echo $(date): "Previous metadata dump found! Removing $FOUND_METADATA"
gcloud storage rm $BUCKET/$FOUND_METADATA
echo $(date): "Previous metadata $FOUND_METADATA removed"
fi
}
function dump_and_cp_metadata() {
echo $(date): "Attempting to dump JuiceFS data"
if [[ -z "$FOUND_METADATA" ]]; then
# If no previous metadata was found, use the current job id
juicefs dump redis://$(echo $WORKER_ADDR):6868/1 $(echo $FLOAT_JOB_ID).meta.json.gz --keep-secret-key
echo $(date): "JuiceFS metadata $FLOAT_JOB_ID.meta.json.gz created. Copying to JuiceFS Bucket"
gcloud storage cp "$(echo $FLOAT_JOB_ID).meta.json.gz" $BUCKET
else
# If previous metadata was found, use the id of the previous metadata
metadata_name=$(echo $FOUND_METADATA | awk -F '.meta.json.gz' '{print $1}')
juicefs dump redis://$(echo $WORKER_ADDR):6868/1 $(echo $metadata_name).meta.json.gz --keep-secret-key
echo $(date): "JuiceFS metadata $metadata_name).meta.json.gz created. Copying to JuiceFS Bucket"
gcloud storage cp "$(echo $metadata_name).meta.json.gz" $BUCKET
fi
echo $(date): "Copying to JuiceFS Bucket complete! Exiting..."
}
# Variables
FOUND_METADATA=""
export GOOGLE_APPLICATION_CREDENTIALS="/mnt/jfs/gcp_cred.json"
# Set Opcenter credentials
install_float
opcenter_ip_address=$(get_secret OPCENTER_IP_ADDRESS)
opcenter_username=$(get_secret OPCENTER_USERNAME)
opcenter_password=$(get_secret OPCENTER_PASSWORD)
# Append to config file
cat <<EOT >> mmc.config
// OpCenter connection settings.
float {
address = '${opcenter_ip_address}'
username = '${opcenter_username}'
password = '${opcenter_password}'
}
EOT
# Initialize gcloud login
gcloud_init
# Start Nextflow run
$nextflow_command
# Error handling - JuiceFS dump and copy to gs bucket
if [[ $? -ne 0 ]]; then
echo $(date): "Nextflow command failed."
remove_old_metadata
dump_and_cp_metadata
exit 1
else
echo $(date): "Nextflow command succeeded."
remove_old_metadata
dump_and_cp_metadata
exit 0
fiJob-Submit Script Adjustments
- Modify the
process.extrawithin themmc.configsection to customize thevmPolicyfor individual nextflow processes (Default policy isspotOnlyin the job submit script. Adjust as needed e.g.,onDemand,spotFirst). You may find more options when callingfloat submit -h:
--vmPolicy [spotOnly=true,retryLimit=10,retryInterval=300s]For handling nextflow code repository, samplesheets and params-file, you have two options: download them or create them directly in the script. Here’s how to do both:
Downloading Samplesheet and Params File
- Provide download commands for users to obtain samplesheet and params file for Nextflow, ensuring you replace
<download-link>with the actual URLs:
# Download samplesheet
gcloud storage cp gs://nextflow-input/samplesheet.csv .
# Download params file
gcloud storage cp gs://nextflow-input/params.yml .Creating Samplesheet and Params File Directly in the Script
- Alternatively, users can create these files directly within the script using the
catcommand as shown below.
Expand to include instructions on creating samplesheet and params files directly in the script
# Create samplesheet
cat > samplesheet.csv << EOF
sample,fastq_1,fastq_2
SAMPLE1_PE,gs://mmc-gcp-poc/viralrecon-test-data/sample1_R1.fastq.gz,gs://mmc-gcp-poc/viralrecon-test-data/sample1_R2.fastq.gz
SAMPLE2_PE,gs://mmc-gcp-poc/viralrecon-test-data/sample2_R1.fastq.gz,gs://mmc-gcp-poc/viralrecon-test-data/sample2_R2.fastq.gz
SAMPLE3_SE,gs://mmc-gcp-poc/viralrecon-test-data/sample1_R1.fastq.gz,
SAMPLE3_SE,gs://mmc-gcp-poc/viralrecon-test-data/sample2_R1.fastq.gz,
EOF
# Create params file
cat > params.yml << EOF
multiqc_title: "rnaseq_multiqc"
fasta: "gs://nextflow-input/reference/Caenorhabditis_elegans.WBcel235.dna.toplevel.fa.gz"
gtf: "gs://nextflow-input/reference/Caenorhabditis_elegans.WBcel235.111.gtf.gz"
save_reference: true
remove_ribo_rna: true
skip_alignment: true
pseudo_aligner: "salmon"
EOF-
Download your nextflow code repository using
git cloneor copying from GCS -
Finally, ensure you customize your
nextflow_commandwith specific pipeline requirements and save the changes:
nextflow_command='nextflow run your_pipeline.nf \
-r your_revision \
-c mmc.config \
-params-file params.yml \
--input samplesheet.csv \
--outdir your_output_directory \
-resume'Remember to replace placeholders with your specific pipeline details.
Additionally, the gs path needs to contain at least a sub-directory and the bucket name MUST NOT contain underscores due to an internal GCS limitation.
Float Submit
- Login to your MMCloud opcenter:
float login -a <opcenter-ip-address> -u <user>- Ensure you are using the latest version of OpCenter & float CLI:
float release upgrade --sync- Make sure you have the following variables set as-is for
float secret's:
+---------------------+
| NAME |
+---------------------+
| OPCENTER_USERNAME |
| OPCENTER_IP_ADDRESS |
| OPCENTER_PASSWORD |
| GCP_BUCKET_JSON |
+---------------------+- You can set the GCP Credential key JSON as secret as below:
float secret set GCP_BUCKET_JSON "$(< gcp_key.json)"- Useful
float secretcommands:
# to list stored secrets
float secret ls
# to set a secret
float secret set OPCENTER_IP_ADDRESS 192.0.1.2
# to unset a secret
float secret unset OPCENTER_IP_ADDRESSFloat Submit Command
- Replace the placeholders
<work-bucket>,<job-name>with your specific values and execute the float submit command:
float submit --hostInit transient_JFS_GCP.sh \
-i docker.io/memverge/juiceflow \
--vmPolicy '[onDemand=true]' \
--migratePolicy '[disable=true]' \
--dataVolume '[size=60]:/mnt/jfs_cache' \
--dirMap /mnt/jfs:/mnt/jfs \
-c 2 -m 4 \
-n <job-name> \
--env BUCKET=gs://<work-bucket> \
-j job_submit_GCP.shPlease Remember to provide the name of your GCP credential key in-between the parentheses.
Here's a brief explanation of the parameters used in the float submit command:
| Parameter | Brief Description |
|---|---|
--hostInit transient_JFS_GCP.sh |
Shell script to run on the host before the job starts. |
-i docker.io/memverge/juiceflow |
Docker image for the job's software environment. |
--vmPolicy '[onDemand=true]' |
Uses on-demand VM instance for head-node execution. |
--migratePolicy '[disable=true]' |
Disables head-node migration to different hosts/VMs. |
--dataVolume '[size=60]:/mnt/jfs_cache' |
Attaches a 60GB data volume at /mnt/jfs_cache in the container. |
--dirMap /mnt/jfs:/mnt/jfs |
Maps a host directory to a container directory for data sharing. |
-c 2 |
Allocates 2 CPU cores to the job. |
-m 4 |
Allocates 4GB of memory to the job. |
-n <job-name> |
Assigns a name to the job for identification. |
--env BUCKET=gs://<work-bucket> |
Sets an environment variable for the GCS bucket URL. |
-j job_submit_GCP.sh |
Specifies the job script or command to run inside the container. |
Important Considerations
-
JuiceFS Bucket Requirements: JuiceFS requires formatting at the root level of a storage bucket. It cannot be formatted on a sub-directory within a bucket. Ensure the root directory of the bucket, or the entire bucket, is specified in the command line interface (CLI) command.
-
Nextflow
-resumeFunctionality: JuiceFlow supports the-resumeoption with Nextflow. Each workflow execution generates a<JOB_ID>.meta.json.gzfile in the work bucket. This file is critical for restoring the work directory for subsequent attempts.
Monitoring on OpCenter
Workflow Execution Log
Once the head node job starts executing, you can monitor the Nextflow standard output in OpCenter GUI as follows:
- Proceed to the OpCenter
Jobsdashboard to monitor the progress of the job

- Once the job starts
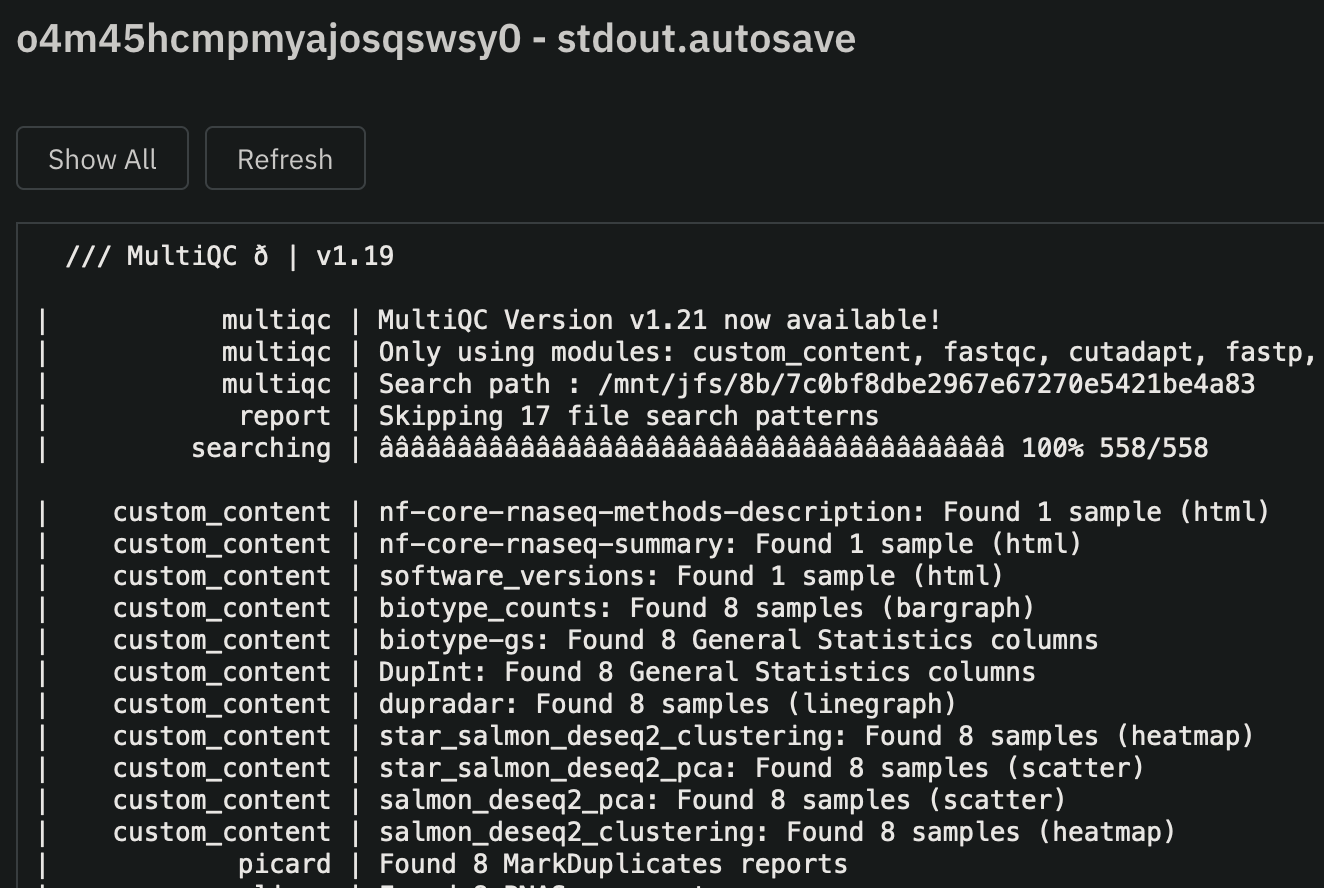
Executing, you can monitor the Nextflow stdout by clicking on the job ->Attachments->stdout.autosave:

To monitor workflow execution and get a detailed view of each process in the Nextflow workflow:
- Click on the
Workflowsdashboard in the OpCenter to monitor workflow execution and get a detailed view for each process in the Nextflow workflow:

- Click on the workflow name, and you can monitor the jobs running in this workflow in this consolidated view:

Individual Job Logs
To view the execution log of any particular job:
- Click on the job-ID.
- Navigate to the
Attachmentstab. - Click
view(eye-icon) on thestdout.autosavefile.
Resuming Workflows with the Job-Submit Script
The job-submit script also allows the resuming of failed Nextflow workflows. You may simply resubmit your previous command again, using the same Bucket. This time, ensure that your Nextflow command include a -resume.
Create Job Templates to Launch via MMCLoud GUI
Job Templates allow for the ease and customaization of runs that follow a similar format, without having the need to manually set up a command every time. It requires the submission of one job first.
- From the
Jobsdashboard, select the jfs-service job previously submitted above and click onMore Actions->Save as Template(NOTE: this is still a BETA feature):

- Provide a name and tag for the private template:

- Navigate to the
Job Templatesdashboard and click onPrivatetemplates:

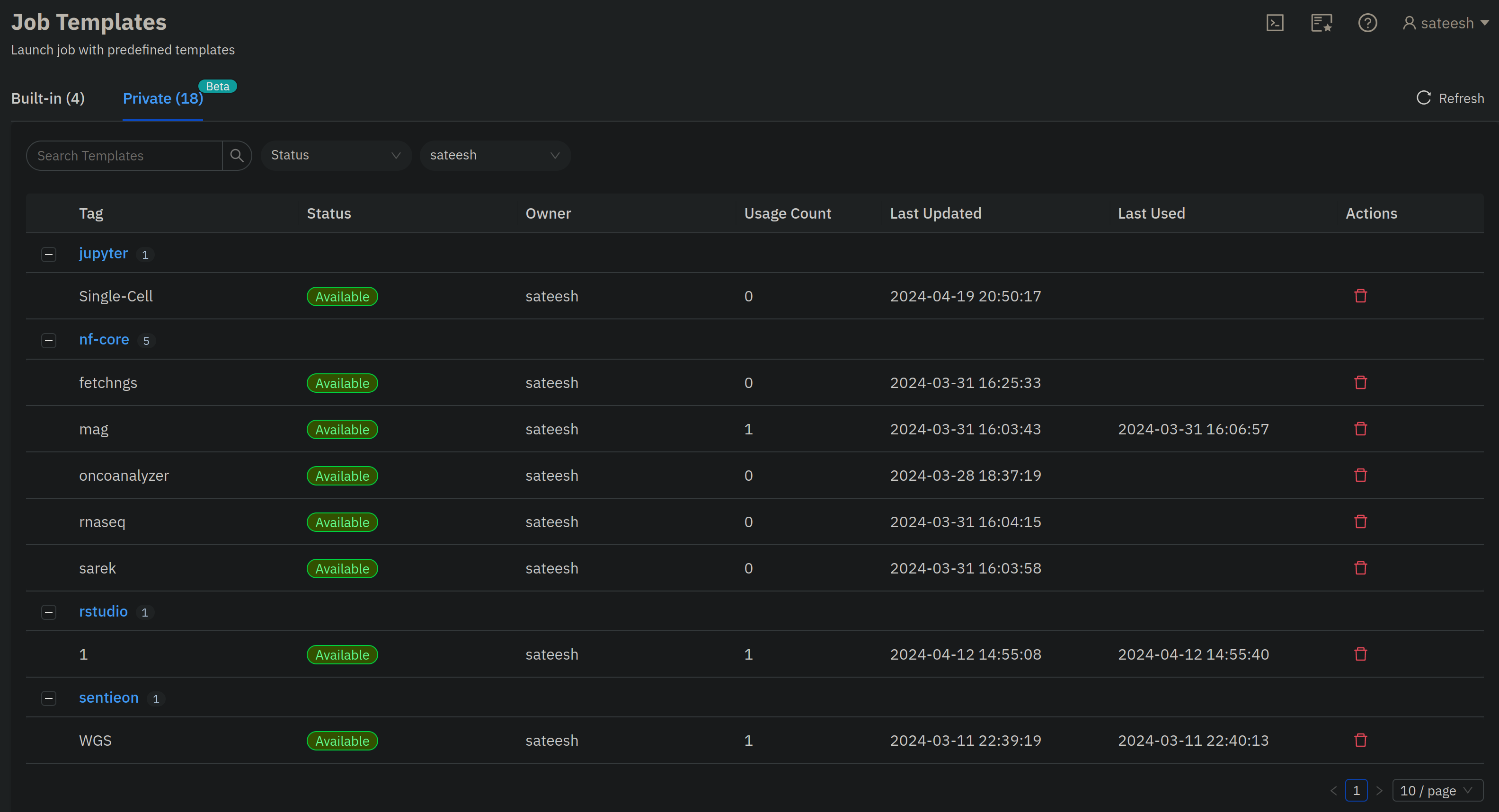
- You can click on any job template, edit/change samplesheet, variables etc., and submit new jobs from the GUI.

Users can also submit jobs from templates via CLI
float submit --template private::<template-name>:<template-tag> \ -e BUCKET=gs://<work-bucket>
Addtionally, please keep in mind the features that need to be updated with every run if they deviate from the default values provided in the private template. This will mainly include:
- GCS Bucket URL
- Updating of the nextflow run command in the job script
