
Sentieon Genomics Tools: Boosting Performance with Memory Machine Cloud (complete report)
Summary
Sentieon® provides a suite of bioinformatics secondary analysis tools that process genomics data with a focus on efficiency, accuracy, and consistency. Compared with comparable open-source software, the Sentieon Genomics tools show a five- to ten-fold improvement in performance.
MemVerge’s Memory Machine Cloud is a software platform that streamlines the deployment of containerized applications in the cloud. Memory Machine Cloud includes a checkpoint/restore feature (called WaveRider) that can automatically migrate a running container, without losing execution state, so that the resources available to a job are never over- or under-provisioned.
When Sentieon Genomics tools are used in conjunction with WaveRider, performance improves even further while costs are also reduced. Depending on how EC2 On-demand instance prices scale with the number of vCPUs and memory capacity, it is possible to optimize cost and performance simultaneously by running a larger virtual machine for a shorter period when CPU and/or memory utilization are high, and a smaller virtual machine for a longer period when CPU and/or memory utilization are low. For example, compared to a baseline measured by running a Whole Genome Sequencing benchmark on a single AWS EC2 instance, the combination of Sentieon Genomics tools and Memory Machine Cloud (with WaveRider) shows a 40% decrease in wall clock time and a 34% reduction in cost.
Introduction
Open-source tools have made analyses of next-generation sequence data practical for a wide array of bioinformaticians. As sequencing machines improve and costs decrease, data sets and data repositories grow larger, emphasizing the need for high-performance software and algorithms. The Sentieon Genomics tools provide a complete rewrite — stressing computational efficiency, accuracy, and consistency — of popular pipelines (such as GATK, Picard, and MuTect) for calling variants from next-generation sequence data. Consistently, Sentieon Genomics tools show a five- to ten-fold improvement in performance over the equivalent open-source pipelines.
Memory Machine Cloud (MMCloud) is a software platform for streamlining the deployment of containerized applications in public clouds such as AWS. A common characteristic of variant calling pipelines is that the resource demands on CPU cores and memory access vary as the pipeline executes. MMCloud includes a checkpoint/restore feature (called WaveRider) that automatically migrates a running container, without losing execution state, to a virtual machine of different capacity as soon as a change in resource requirements is detected.
When used with WaveRider, Sentieon Genomics tools execute faster (measured by wall clock time) because processes are never starved for resources. The costs associated with pipeline execution also decrease because virtual machines are never over-provisioned.
This report compares the performance of a whole genome sequencing (WGS) pipeline benchmark (developed by Sentieon) executed without using MMCloud to the performance when used with MMCloud.
WGS Pipeline Benchmark
The Sentieon-developed WGS pipeline benchmark simulates a typical bioinformatic analysis pipeline, starting with loading raw data (in the form of fastq files) and ending with output in the form of vcf files. The WGS pipeline benchmark comprises five steps as shown in the table.
| Step | Function | Description | Output | Example of open-source analogue |
| 1 | Data input | Dataset downloaded from repository (such as AWS S3) and loaded into local storage | Dataset available on local disk | NA |
| 2 | Sequence mapping | Align local sequence to reference genome | Coordinate-sorted bam file | BWA-MEM |
| 3 | De-duplication | Remove duplicate reads | De-duplicated bam file | Samtools or Picard (included in GATK) |
| 4 | Quality Control | BQSR (Base Quality Score Recalibration) | Recalibrated base scores in new bam file | GATK |
| 5 | Variant calling | Haplotype-aware variant detection | Variant Call Format (VCF) file | GATK |
Performance Analysis
The Sentieon Genomics tools are used in a single-server application, that is, a single script configures the tools covering multiple data processing steps on one machine. A third-party workflow management application is not needed to drive the pipeline. This makes comparative performance analysis unambiguous — any performance differences cannot be attributed to the workflow manager.
For the performance analysis reported here, the WGS pipeline is segmented into three phases (numbered steps refer to the previous table).
- Phase 1: Data Input (Step 1)
- Phase 2: Sequence Mapping (Step 2)
- Phase 3: GATK procedures (Steps 3 to 5)
To establish a baseline, the WGS pipeline benchmark is run on an On-demand virtual machine instance in the Amazon AWS cloud. One instance of an r5.8xlarge (32 vCPUs and 256 GB memory capacity) is provisioned and this single instance is used for the entire pipeline. Although 128 GB is sufficient memory capacity for this pipeline, the r5.8xlarge instance was selected over a smaller virtual machine (16 vCPUs and 128 GB memory capacity) because the r5.8xlarge instance was available at a lower price. This means that the WGS pipeline benchmark runs can be used as a performance baseline as well as a cost baseline.
The prices quoted here are the prices published by AWS at the time the tests were conducted. Prices fluctuate according to market conditions.
WGS Pipeline Benchmark Baseline
To determine the baseline metrics to compare with later tests, the WGS Pipeline Benchmark was run on a single r5.8xlarge instance. The graphical display obtained from the MMCloud web interface is shown in Figure 1. The display of CPU utilization (summed over all vCPUs so the maximum possible is 3200%) is annotated to show the wall clock times corresponding to Phase 1, Phase 2, and Phase 3, respectively. Numerical results are summarized in Table 1.
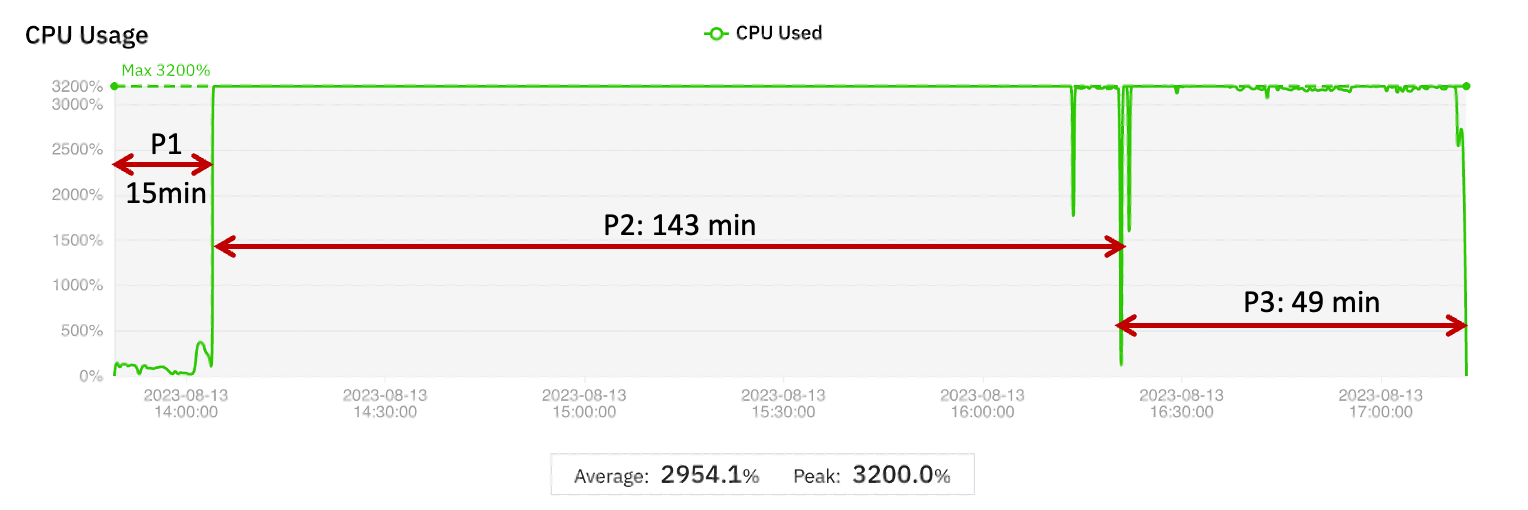
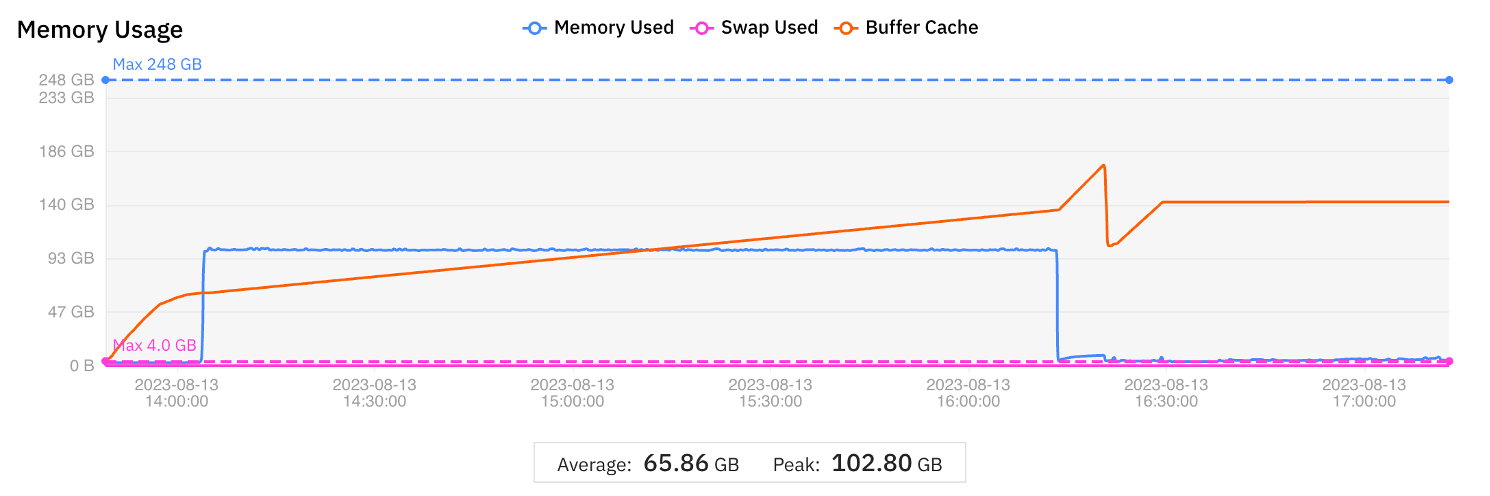
Figure 1 Graphical display of CPU and Memory utilization for the WGS Pipeline Benchmark test to establish baseline metrics
| Phase 1 | Phase 2 | Phase 3 | Complete run | |
| VM instance | r5.8xlarge | r5.8xlarge | r5.8xlarge | |
| vCPUs | 32 | 32 | 32 | |
| Memory (GB) | 256 | 256 | 256 | |
| Wall clock time (min) | 15 | 143 | 49 | 207 |
| Cost (USD) | 0.51 | 4.87 | 1.67 | 7.06 |
Table 1 Wall clock time and cost for the WGS Pipeline Benchmark test
Discussion
The WGS Pipeline Benchmark run shows three distinct phases.
- Phase 1 only uses one vCPU – the CPU utilization is 100% or less compared to a theoretical maximum of 3200%. Memory utilization is also low.
- Phase 2 fully utilizes all thirty-two vCPUs by using multi-threaded processing. Memory utilization is much higher than in Phase 1, although the peak (103 GB) is still much less than the 256 GB capacity of the virtual machine instance.
- Phase 3 also uses all thirty-two vCPUs but the memory utilization is low (comparable to Phase 1).
If the WGS Pipeline Benchmark is representative of a large class of pipelines, then the CPU and memory utilization profiles indicate areas where performance and cost can be optimized. The ratio of peak memory usage to average memory is large, which means that for a significant part of the run, memory capacity is overprovisioned even when CPU utilization is high. If the virtual machine is sized to the average memory used, then the job would either run slowly (because of excessive memory swapping) or fail due to out-of-memory (OOM) errors.
WGS Pipeline Benchmark with MMCloud
MMCloud’s WaveRider feature migrates a running job, without losing state, from one virtual machine to another. The migration can be initiated manually (using the CLI or the web interface), programmatically (inserting CLI commands into the job script), or by policy (crossing CPU and/or memory utilization thresholds). The ability to migrate a running job means that, as the job runs, the underlying virtual machine can be sized appropriately for the execution phase.
To investigate the effect of WaveRider on WGS Pipeline Benchmark metrics, migration events were triggered programmatically at the junctions where one phase ends and a new phase starts.
Cost Optimization
To optimize cost, the following test was conducted. The CLI commands are displayed in the Appendix.
- Start Phase 1 with a small virtual machine (4 vCPUs and 32 GB memory)
- At the start of Phase 2, migrate to a large virtual machine (32 vCPUs and 256 GB memory)
- At the start of Phase 3, migrate to a virtual machine with the same number of vCPUs but smaller memory capacity (64 GB)
The results are displayed graphically in Figure 2. The time taken to migrate to a new virtual machine is included in the wall clock time.
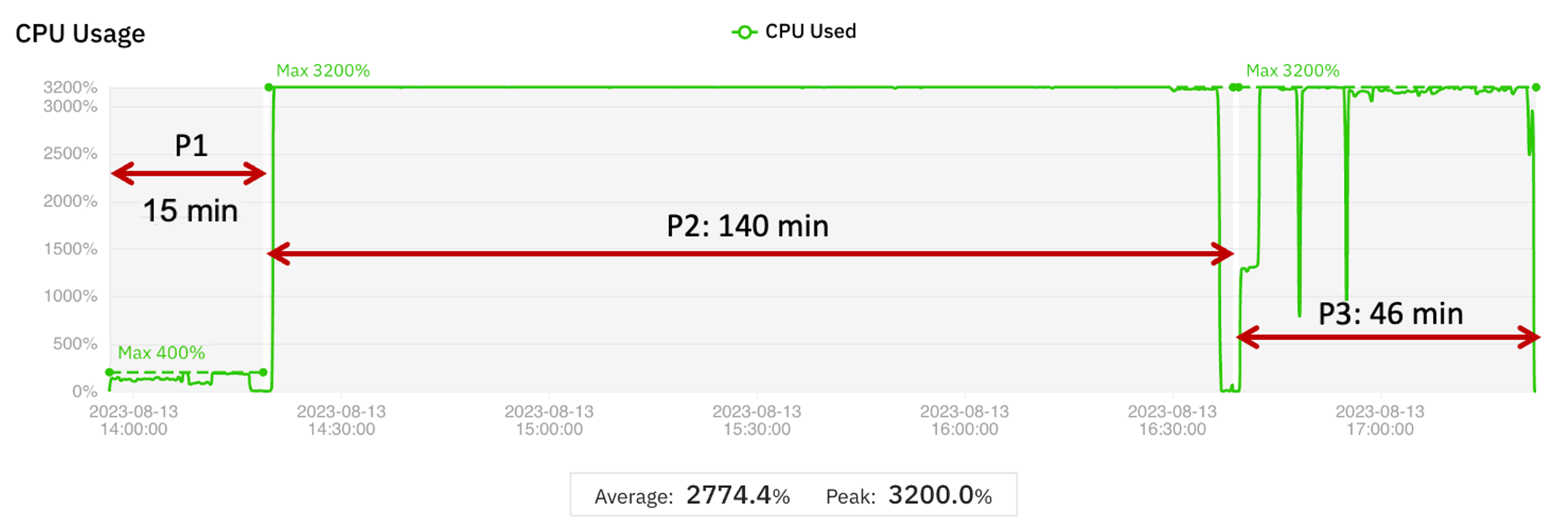
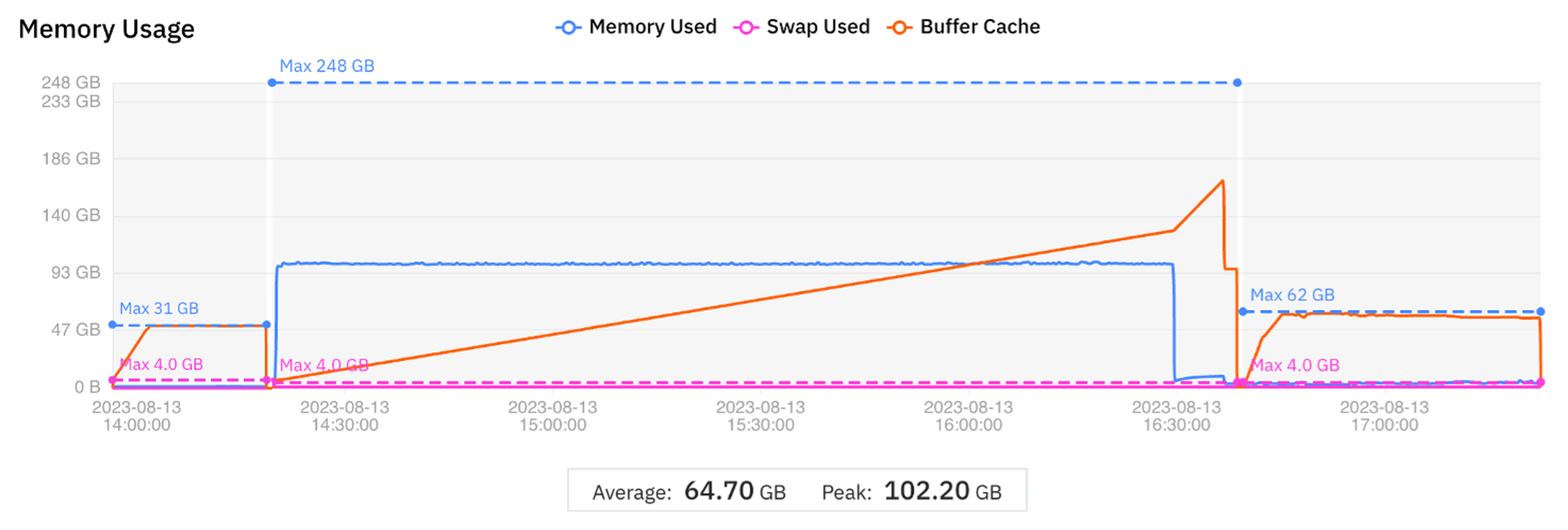
Figure 2 Graphical display of job migration to optimize cost
The comparison with the baseline is shown in Table 2.
| Phase 1 | Phase 2 | Phase 3 | Complete run | |
| VM instance | r5.xlarge | r5.8xlarge | c6i.8xlarge | |
| vCPUs | 4 | 32 | 32 | |
| Memory (GB) | 32 | 256 | 64 | |
| Wall clock time (min) | 15 | 140 | 46 | 201 |
| Difference from baseline (%) | 0 | -2 | -6.1 | -2.9 |
| Cost (USD) | 0.09 | 4.79 | 1.04 | 5.92 |
| Difference from baseline (%) | -82.4 | -1.7 | -38.0 | -16.1 |
Table 2 Comparison of cost optimization test with baseline metrics
Discussion
Although the goal of this test is to optimize cost (the results show a 16% decrease), the overall wall clock time also decreases slightly. The wall clock times for Phases 2 and 3 are within the expected variation and may be considered the same as the comparable phases in the baseline run.
As the size of the dataset grows, the time that Phase 1 takes to complete can become larger than the time to complete Phases 2 and 3. In this case, cost savings increase even more (compared to the baseline) if a small virtual machine is used for Phase 1. An example is shown in Figure 3.
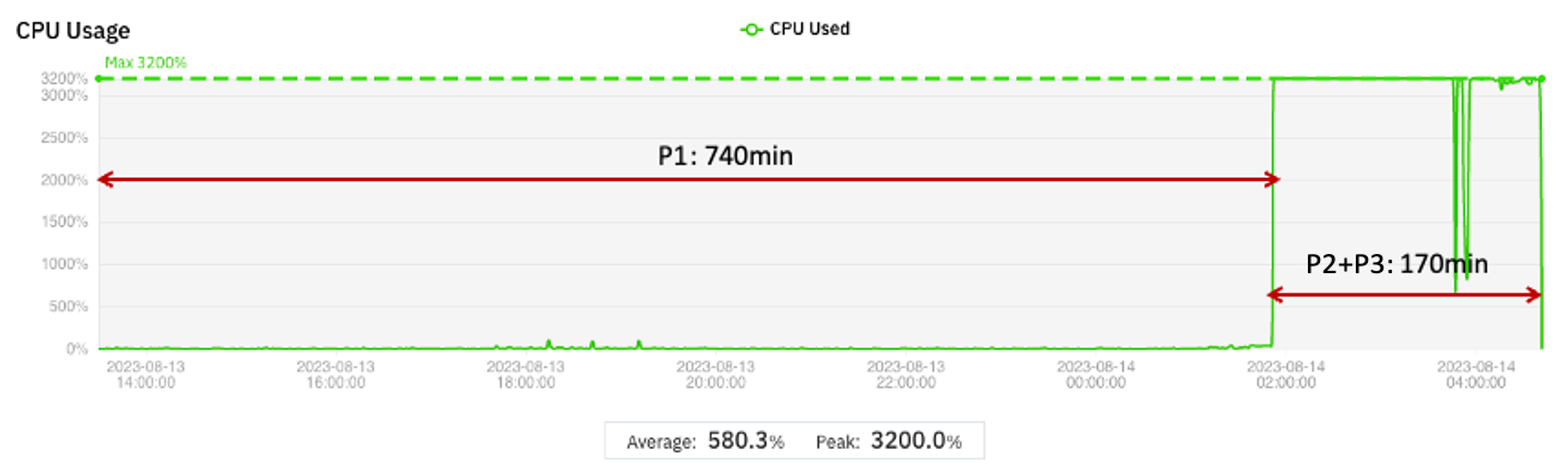
Figure 3 Cost optimization for extended Phase 1 case
If a t3.medium instance is used for Phase 1 and an r5.8xlarge instance is used for Phase 2, the estimated cost savings for a pipeline with this profile are 79% compared to running this pipeline entirely on a r5.8xlarge instance.
Cost and Performance Optimization
A key component in optimizing software performance is to ensure that processes are not delayed because they are waiting for resources to free up. The Sentieon Genomics tools provide a configuration option to specify the maximum number of process threads. In this test, the maximum number of threads is increased to 64 because the number of vCPUs for the Phase 2 virtual machine is increased to 64. If the maximum thread of threads is greater than the number of vCPUs, the effect on performance is neutral or negative (because of excessive context switching). For this reason, the baseline uses 32 as the maximum number of threads.
To optimize performance, the following test was conducted. The CLI commands are displayed in the Appendix.
- Increase the maximum number of threads to 64
- Start Phase 1 with a small virtual machine (4 vCPUs and 32 GB memory)
- At the start of Phase 2, migrate to a large virtual machine (64 vCPUs and 256 GB memory)
- At the start of Phase 3, decrease the maximum number of threads to 32 and migrate to a virtual machine with 32 vCPUs and 64 GB memory
The results are displayed graphically in Figure 4. The time taken to migrate to a new virtual machine is included in the wall clock time
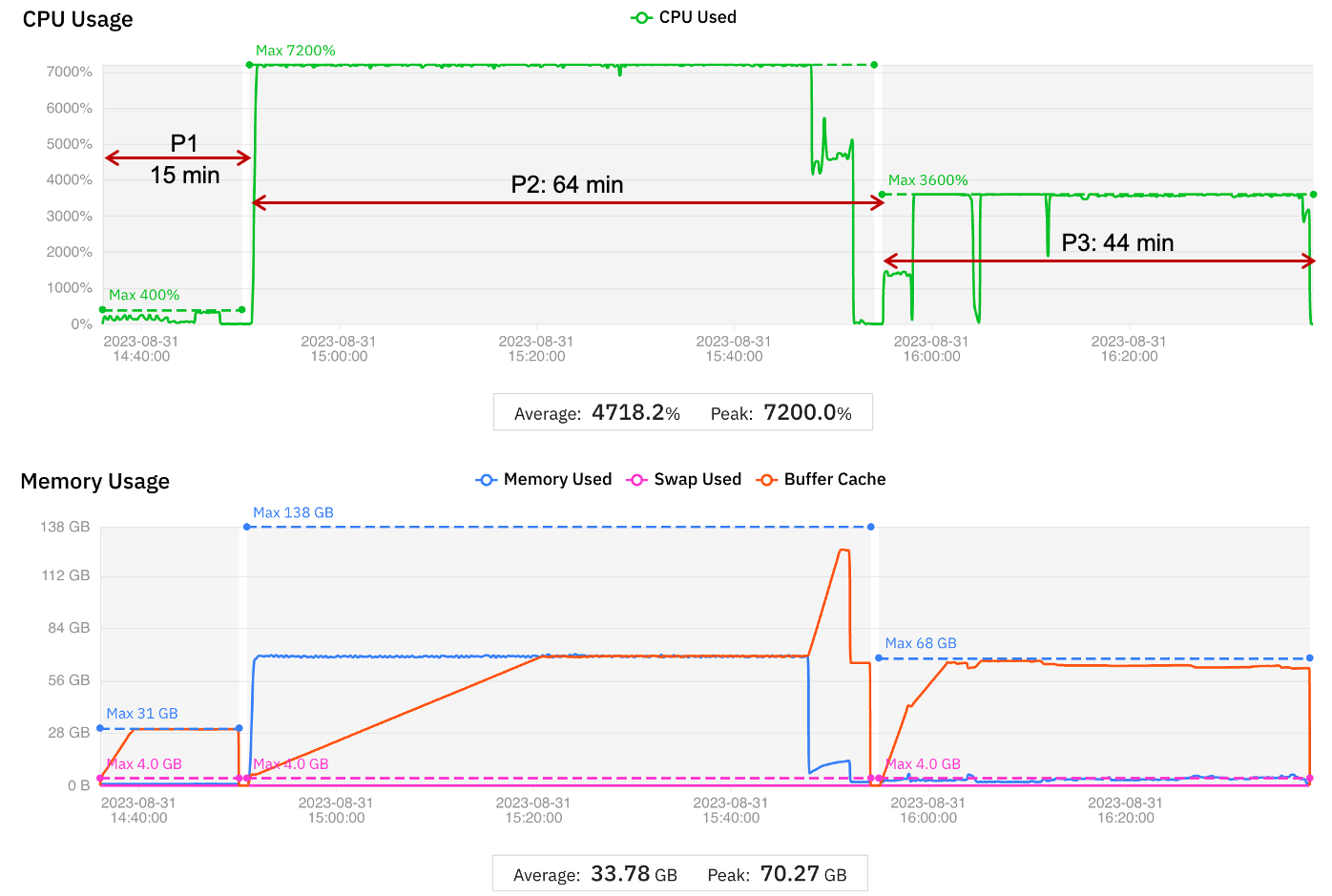 Figure 4 Graphical display of job migration to optimize performance
Figure 4 Graphical display of job migration to optimize performance
The comparison with the baseline is shown in Table 3.
| Phase 1 | Phase 2 | Phase 3 | Complete run | |
| VM instance | r5.xlarge | c5.18xlarge | c5.9xlarge | |
| vCPUs | 4 | 72 | 36 | |
| Memory | 32 | 144 | 72 | |
| Running Time (min) | 15 | 64 | 44 | 123 |
| Difference from baseline (%) | 0 | -55.2 | -10.2 | -40.5 |
| Cost (USD) | 0.09 | 3.41 | 1.19 | 4.69 |
| Difference from baseline (%) | -82.4 | -30.0 | -28.7 | -33.6 |
Table 3 Comparison of performance optimization test with baseline metrics
Discussion
There are two options for specifying the virtual machine size when submitting a job to MMCloud:
- Specify the instance type by name, for example, r5.8xlarge is an EC2 instance type in AWS, or
- Specify a range of vCPUs and range of memory capacity, for example, the number of vCPUs (memory) must be at least 32 (128 GB)
With the second option, MMCloud selects the virtual machine instance with the lowest price that meets the vCPU and memory requirements. Sometimes a larger virtual machine is available at a lower price and sometimes a virtual machine with the minimum requirements is not available. For example, in this test MMCloud searched for a virtual machine with at least 64 vCPUs and 128 GB and found a virtual machine with 72 vCPUs and 144 GB memory. With the more capable virtual machine, Phase 2 completed in less than half the wall clock time while still reducing Phase 2 cost by 30% (compared to the baseline).
Conclusion
Sentieon Genomics tools decrease wall clock times for executing a wide range of bioinformatic pipelines. Benchmark tests show that, by using MMCloud’s WaveRider feature to rightsize compute instances, wall clock time can be reduced by a further 40%. Moreover, the performance improvement is accompanied by significant cost savings (34%) because the Sentieon software can take advantage of extra vCPUs — the more capable virtual machine decreases wall clock time faster than the cost per unit time increases for additional vCPUs. Cost savings also depend on the relative time spent loading data versus computation.
Additional cost savings can be realized by using Spot instance instead of On-demand instances and using MMCloud’s checkpoint/restore feature to handle any Spot instance reclaim events without losing state. This aspect is not investigated in this report.
Appendix: MMCloud CLI Commands
MMCloud includes a CLI called float that is used to schedule and manage pipelines. This appendix contains the CLI commands used in the tests.
Cost Optimization
The following procedure was used to schedule the WGS Pipeline Benchmark.
- Schedule pipeline using the float submit command with the
-c 4 -m 8options. - Insert the following float command before the start of Phase 2 to migrate the job to a compute instance with at least 32 vCPU and 128 GB memory.
/opt/memverge/bin/float migrate --sync -f -c 32 -m 128 -j ${FLOAT_JOB_ID}
- Insert the following float command before the start of Phase 3 to migrate the job to a compute instance with at least 32 vCPUs and 64 GB memory.
/opt/memverge/bin/float migrate --sync -f -c 32 -m 64 -j ${FLOAT_JOB_ID}
Cost and Performance Optimization
This test increases th enumber of CPU cores for Phase 2 to optimize performance (64 vCPUs or double the number of vCPUs used in the baseline test). The following procedure was used to schedule the WGS Pipeline Benchmark.
- Change the maximum number of threads from 32 to 64 by setting
nt=64or using the-t 64option in the sentieon command string. - Schedule pipeline using the float submit command with the
-c 4 -m 8options. - Insert the following float command before the start of Phase 2 to migrate the job to a compute instance with at least 64 vCPUs and 128 GB memory.
/opt/memverge/bin/float migrate --sync -f -c 64 -m 128 -j ${FLOAT_JOB_ID}
- Before the start of Phase 3, change the maximum number of threads to 32 (set
nt=32), and insert the following float command to migrate to an instance with 32 vCPUs and 64 GB memory.
/opt/memverge/bin/float migrate --sync -f -c 32 -m 64 -j ${FLOAT_JOB_ID}